As new research led by Oncode Investigator Tineke Lenstra (The Netherlands Cancer Institute) shows, our cells still contain many secrets. Together with her team, Lenstra developed an algorithm that can track gene expression in 3D over time. This allowed them to create incredibly detailed recordings of what happens inside a cell. As it turns out, there’s somewhat of a relay race happening on our DNA, allowing cells to produce the necessary proteins. This insight and discovery didn’t escape the renowned microscopy company Zeiss. Through a collaboration facilitated by Oncode Institute, Zeiss licensed the algorithm and will be releasing it making it available to all researchers across fields. Moreover, the algorithm and new insights from this study were recently published in Molecular Cell.
Deregulation of transcription factors (TF) is critical to hallmarks of cancer. Genetic mutations, gene fusions, amplifications or deletions, epigenetic alternations and aberrant posttranslational modification of TFs are involved in the regulation of various stages of carcinogenesis, including cancer initiation, progression, and metastasis. Thus, understanding the mechanisms of TFs will aid developing novel strategies for dysfunctional TFs. Despite being such a fundamental process, the temporal regulation of the central dogma of gene expression by TFs is still not well understood.
Mini shifts
Oncode Investigator Tineke Lenstra’s research group discovered that TF’s perform somewhat of a relay race while activating gene expression. That allows them to let the DNA be read long enough despite their brief shifts that last mere seconds. Their seamless work allows the cell to create enough protein to function properly.
Text continues below the image.
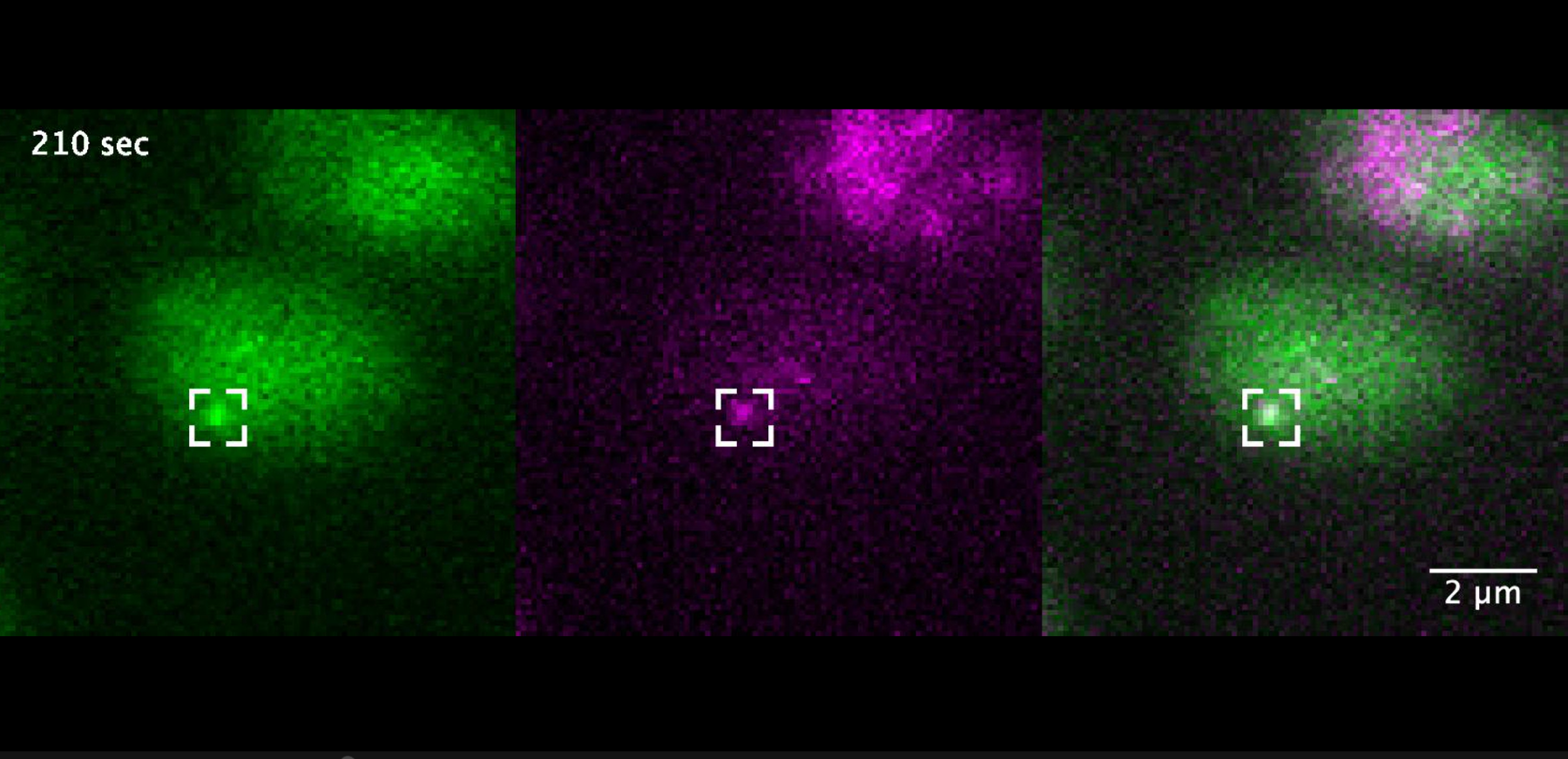
Image above: Green (left) shows transcription of a gene (GAL10) – the brighter the dot, the more the gene is transcribed. Purple measures transcription factors that wander around the cell nucleus or bind to DNA. On the right, these two images are superimposed: purple and green overlap where the transcription factor binds to the gene.
Recording genes
The achievement of Tineke Lenstra and her team in examining living cells with unprecedented detail represents a significant milestone in the field of cellular biology. The inherent challenge in such an endeavor stems from the perpetual movement within the cells, complicating efforts to capture and study these dynamics through microscopy. To overcome this obstacle, the researchers devised a sophisticated algorithm enabling the microscope to meticulously follow a specific gene within the living cells, regardless of its continuous movement. This algorithm employs a method to "pin" a certain point within the microscopy imaging, thus facilitating precise monitoring of transcription factor DNA binding at single-molecule resolution. This technological advancement opens up new vistas for the in-depth exploration of gene expression.
License Deal with Zeiss
Zeiss, a leading company in microscopy, recognized the value of this innovative approach and, with the support of the Oncode Institute, secured a license for the algorithm. This move ensures that the method can benefit a broad spectrum of researchers across various scientific disciplines “It’s great that a method that we developed for a highly fundamental research question can be useful to various researchers from across all sorts of disciplines” says Lenstra.
This research was highlighted in a Mol Cell preview, dive deeper in the technology & impact in the same Mol Cell issue.
For more information on this please contact Tineke Lenstra at [email protected] or Yuva Oz at [email protected].
This research was financially supported by KWF Dutch Cancer Society, Oncode Institute and the European Research Council. The collaboration between Zeiss and Tineke Lenstra and further licensing of the algorithm was facilitated by Oncode Institute.