In this overview, we describe the primeTF methodology: what it can be used for, how it compares to related technologies, and its main advantages and limitations.
Developed by the Bas van Steenstel Group
What is the methodology good for?
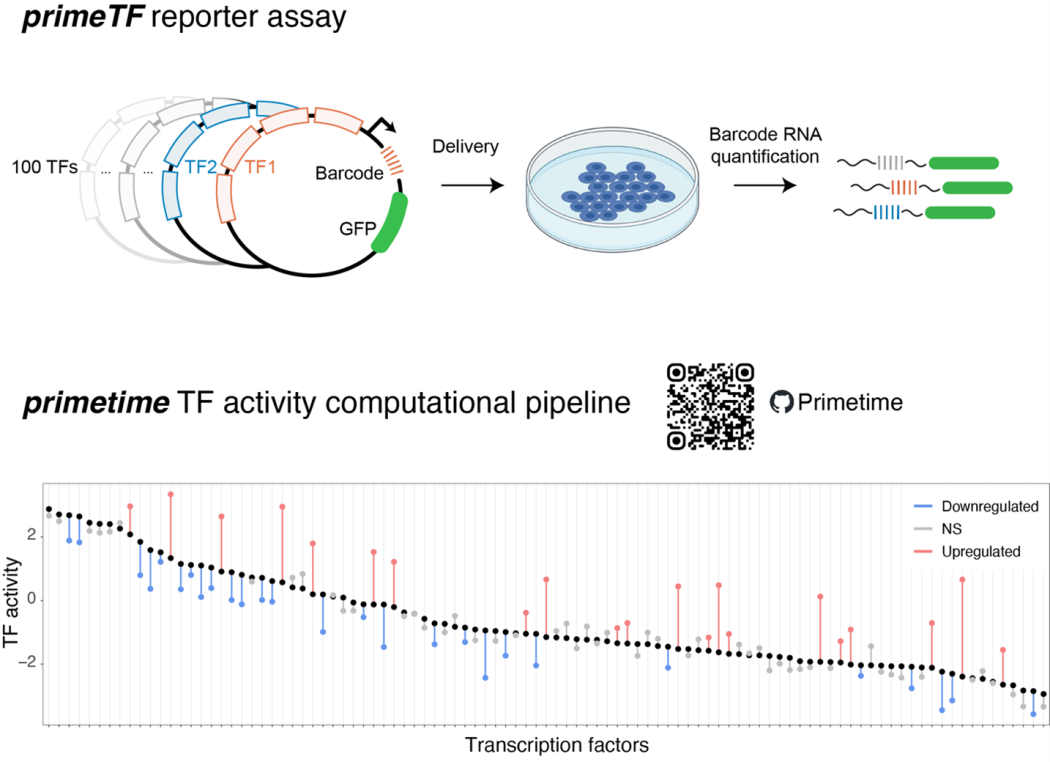
primeTF enables sensitive and multiplexed measurement of transcription factor (TF) activity in cultured cells, using a collection of 100 TF highly optimized reporters that are assembled into a single barcoded reporter library. This library can be delivered into any cell culture model via transfection or lentiviral transduction. TF activities are then determined by barcode counting in RNA. Our automated computational pipeline, primetime, streamlines the analysis and identifies differential TF activity across experimental conditions. Together, the reporter library and pipeline serve as a powerful tool to characterize changes in the “regulatory state” of the cell in response to stimuli, drug treatments and genetic perturbations.
What is/are the main advantages of this methodology over related technologies?
Unlike RNA-seq or ATAC-seq, primeTF directly assesses transcriptional activity, providing higher specificity for TF function. The activity of 100 TFs can be measured in a single tiny dish of cells (for some cell lines in a single well of a 96-well plate). The protocol requires only standard molecular biology reagents and <100,000 Illumina reads per sample, making it affordable and scalable.
What are the most important limitations of the methodology?
The method does not currently support single-cell resolution. Efficient reporter delivery into the cell model of interest is required — models with low delivery efficiency may need higher input cell numbers.
What type of samples are compatible with methodology (yes, no, possibly)?
Cancer cell lines | Primary cells in culture | Organoids | Primary tissue |
Yes | Yes | Yes | No |
What future develops to the methodology are you planning, if any?
We are developing an automated 384-well screening workflow for cost-efficient, high throughput data generation. In parallel, we are implementing nascent RNA readouts using metabolic labeling to increase the temporal resolution of TF activity measurements.
If someone outside your lab wants to use the methodology, what is the best option?
A detailed protocol for both the experimental and computational workflow is available. The primeTF reporter library is available upon signing an MTA. Researchers with access to the library and protocol can perform experiments independently.
Name one or more people in your lab that are experienced with the methodology
Max Trauernicht ([email protected]), Hatiçe Yücel ([email protected])
Who originally developed the methodology?
The optimized TF reporters were developed by Max Trauernicht in the van Steensel group, as published here: https://doi.org/10.1016/j.cels.2024.11.003. The automated workflow and primetime pipeline were developed by Hatice Yücel, Vinícius H. Franceschini-Santos and Max Trauernicht.
Confidentiality Statement
This bulletin contains proprietary information intended solely for the designated recipients. Redistribution, forwarding, copying, or any form of disclosing it to third parties- whether in part or in full - is strictly prohibited without the explicit prior written permission of Oncode Institute. By accessing this material, you agree to comply to these confidentiality obligations and to use it solely for its intended informational purpose.


