In this overview, we describe the PTATO methodology: what it can be used for, how it compares to existing technologies, and its main advantages and limitations.
What is the methodology good for?
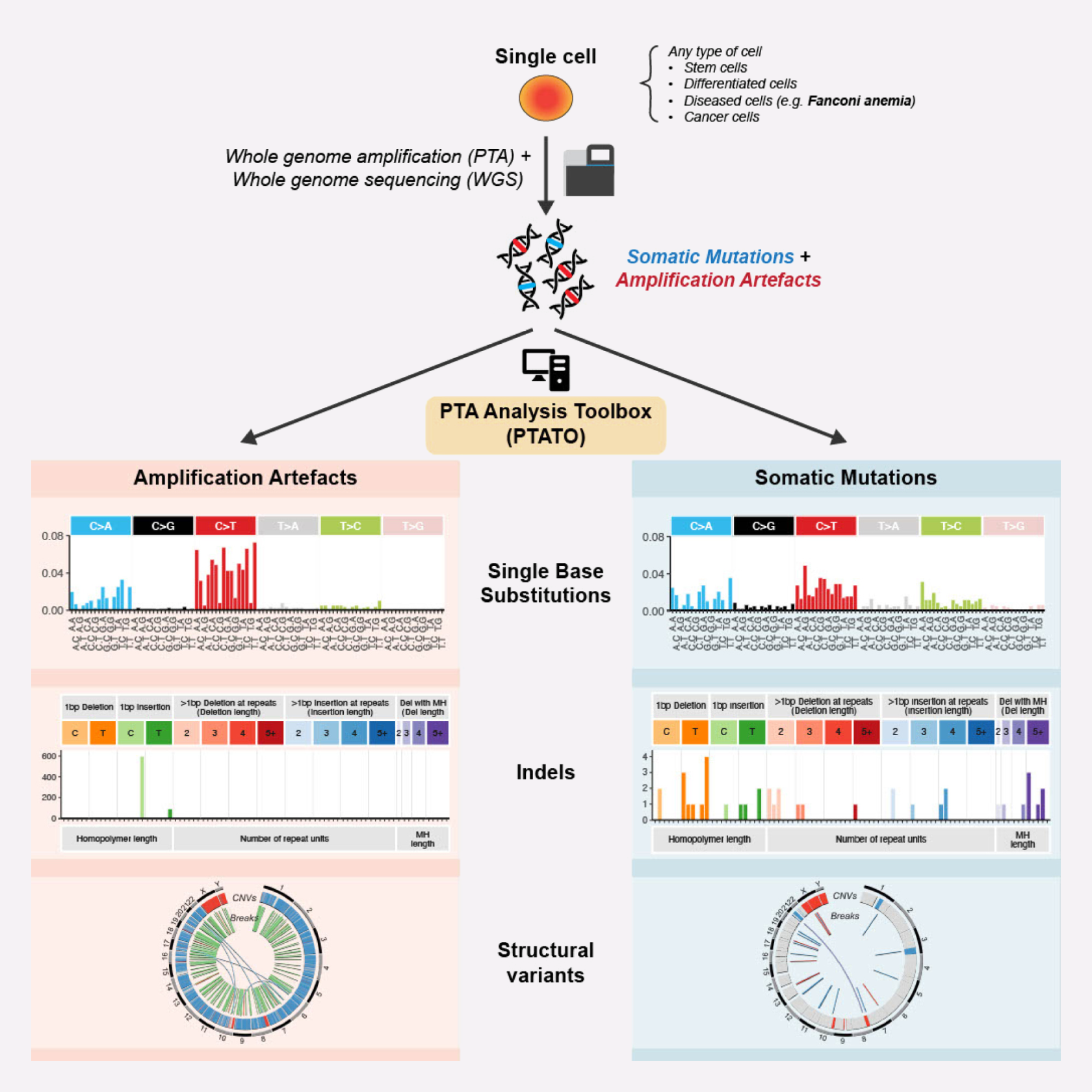
Whereas single-cell genomics tools have revolutionized biomedical research, single-cell whole genome sequencing has lagged because of biases introduced during DNA amplification. Our method allows for effective flagging and filtering of such artifacts allowing for accurate mutation discovery of individual cells at nucleotide resolution. This technology enables to study genetic heterogeneity, retrospectively trace clonal lineages and perform mutational signature analysis in any viable cell.
PTATO: a single-cell genome analysis toolkit for nucleotide resolution mutation discovery
What is/are the main advantages of this methodology over related technologies (1-3 sentences)?
We make use of a random forest model that was trained using unique sequencing data and which can classify individual mutation calls as an artefact or not. Previous approaches made use of standard filtering criteria, which filters a significant proportion of true mutations while also allowing many artefacts to pass the filter. We have benchmarked our approach to the existing state-of-the-art and demonstrate that PTATO is superior.
What are the most important limitations of the methodology (1-3 sentences)?
Single-cell whole genome sequencing is expensive and because of this a limited number of cells per sample can be analyzed (10s – 100s of cells).
What type of samples are compatible with methodology (yes, no, possibly)?
Cancer cell lines | Primary cells in culture | Organoids | Primary tissue |
Yes | Yes | Yes | Yes |
What future develops to the methodology are you planning, in any?
Our method allows for assessing both the genome and transcriptome of the same cell. However, due to RNA contamination in the DNA fraction of the cell, the single-cell whole genome sequencing data is “contaminated” with RNA editing events. We have currently developed an add-on to PTATO that allows for filtering RNA reads from the DNA reads enabling combined DNA/RNA analysis on nucleotide resolution. Of note, the throughput of our method is limited and integration with standard 10X single-cell RNA sequencing data is possible but not optimal.
If someone outside your lab wants to use the methodology, what is the best option?
A) What do you need to provide them to make it work (select one or more)?
Train a person from their lab
B) Is there any minimal expertise/equipment others need to work with the methodology?
Bioinformatics skills are required. Also, the whole genome amplification steps require a bit of training.
Name one or more people in your lab that are experienced with the methodology
Markus van Roosmalen developed, maintains and constantly optimizes PTATO ([email protected]).
Laurianne Trabut has optimized the whole genome amplification procedure ([email protected]).
Who originally developed the methodology?
Our group has originally developed PTATO (Middelkamp et al, Cell Genomics 2023 & Derks*, van Leeuwen* et al, STAR Protocols 2024) and it makes use of a previously developed method by others to amplify the DNA of a single cell, named primary template-directed amplification (Gonzalez-Pena et al, PNAS 2021).
Confidentiality Statement
This bulletin contains proprietary information intended solely for the designated recipients. Redistribution, forwarding, copying, or any form of disclosing it to third parties- whether in part or in full - is strictly prohibited without the explicit prior written permission of Oncode Institute. By accessing this material, you agree to comply to these confidentiality obligations and to use it solely for its intended informational purpose.


